|
|
Journal of Advanced Veterinary Research Volume 9, Issue 2, 2019, Pages: 64-68 |
|
|
PCR Identification of Trypanosomes Isolated from Cattle and Glossina spp. in Wildlife- Human-animal Interface of Meatu District, North-Eastern Tanzania |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Deusdedit J. Malulu1,3, Huruma N. Tuntufye1, Benigni A. Temba2, Elikira N. Kimbita1, Imna. I Malele3, Safari Kinung’hi4, Hamisi S. Nyingilili3, Togolai Mbilu5, Josephat S. Kaboya5, Eugene Lyaruu3 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
1Department of Microbiology, Parasitology and Biotechnology, Sokoine University of Agriculture, Tanzania. 2Department of Veterinary Physiology, Biochemistry and Toxicology, Sokoine University of Agriculture, Tanzania. 3Vector and Vector-Borne Diseases Institute, Tanga, Tanzania. 4National Institute for Medical Research, Mwanza Centre, Tanzania. 5National Institute for Medical Research, Tabora Centre, Tanzania. *Corresponding author: deusdedit.malulu@tvla.go.tz |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Received: 15 March 2019, Accepted: 5 April 2019 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
African trypanosomes are etiological agents of trypanosomosis transmitted by tsetse flies (Glossina spp). Thus, identifying them in vectors and hosts together with their classification into species, subspecies is crucial for effective control of the diseases they cause to animals and human. This study analysed 350 samples collected from cattle (100) and tsetse flies (250) of Meatu district for identification of trypanosomes through amplification of Internal transcribed spacer 1 (ITS-1) region in order to support formulation of tsetse and trypanosomosis control strategies within the district. Occurrence of trypanosomes in cattle was 15 %, while in tsetse was 1.20 %. Trypanosoma congolense was identified in cattle whereas; T. simiae and T. godfreyi were identified in tsetse flies. The tsetse and trypanosomosis control options were discussed. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Keywords: ITS1 PCR, Tanzania, trypanosomes, trypanosomosis, tsetse fly |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Introduction |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Characterization of tsetse transmitted trypanosomes has been a subject of research since late 18th century as agents of African Animal Trypanosomosis termed as “nagana” in cattle, and later Human African Trypanosomosis commonly known as sleeping sickness in humans (Adams et al., 2008; Steverding, 2008). Tsetse borne sleeping sickness and nagana impede livestock sector development in sub-Saharan Africa where thousands of human lives are threatened and potential agricultural areas are underutilized (Ilemobade, 2009). Milestones reached on the characterization of trypanosomes in vectors and vertebrates includes; genus and sub-genus levels discrimination henceforth distinction of African trypanosomes from American trypanosomes, discovery of new species and subspecies and existence of wide range of reservoirs hosts (Lloyd and Johnson, 1924; Hoare, 1972). Moreover, evidences have been made to infer that animal and human infective trypanosomes are different in respect to their pathogenicity and host range (Gibson, 2003). This information has been useful tool in decision making regarding tsetse control. Characterizing human pathogens in relation to animal pathogens is consequently a public health issue and suits better allocation of the limited resources for diseases surveillance and control (Shaw et al., 2014). Specification of the pathogens also facilitates combating sleeping sickness, which is usually expensive to control and highly life threatening (Sindato et al., 2008). Tanzania has seven known species of Glossina, which are infested by Trypanosoma congolense, T. vivax, T. brucei brucei, T. godfreyi and T. simiae that cause nagana in livestock. Trypanosoma brucei rhodesiense is currently reported sole cause of sleeping sickness in human (Kibona, 2001; Daffa et al., 2013). There was a need to generate enough information regarding sleeping sickness in Meatu district. The sleeping sickness cases reported in recent years and the use of trypanocides among livestock keepers (District unpublished report, 2015) influenced this study to be conducted. The fact that the district has potential for both tourism and animal husbandry, which offer livelihood to majority households, revealing sleeping sickness and nagana status could support strategic tsetse control in the district given limited resources. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Materials and methods |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
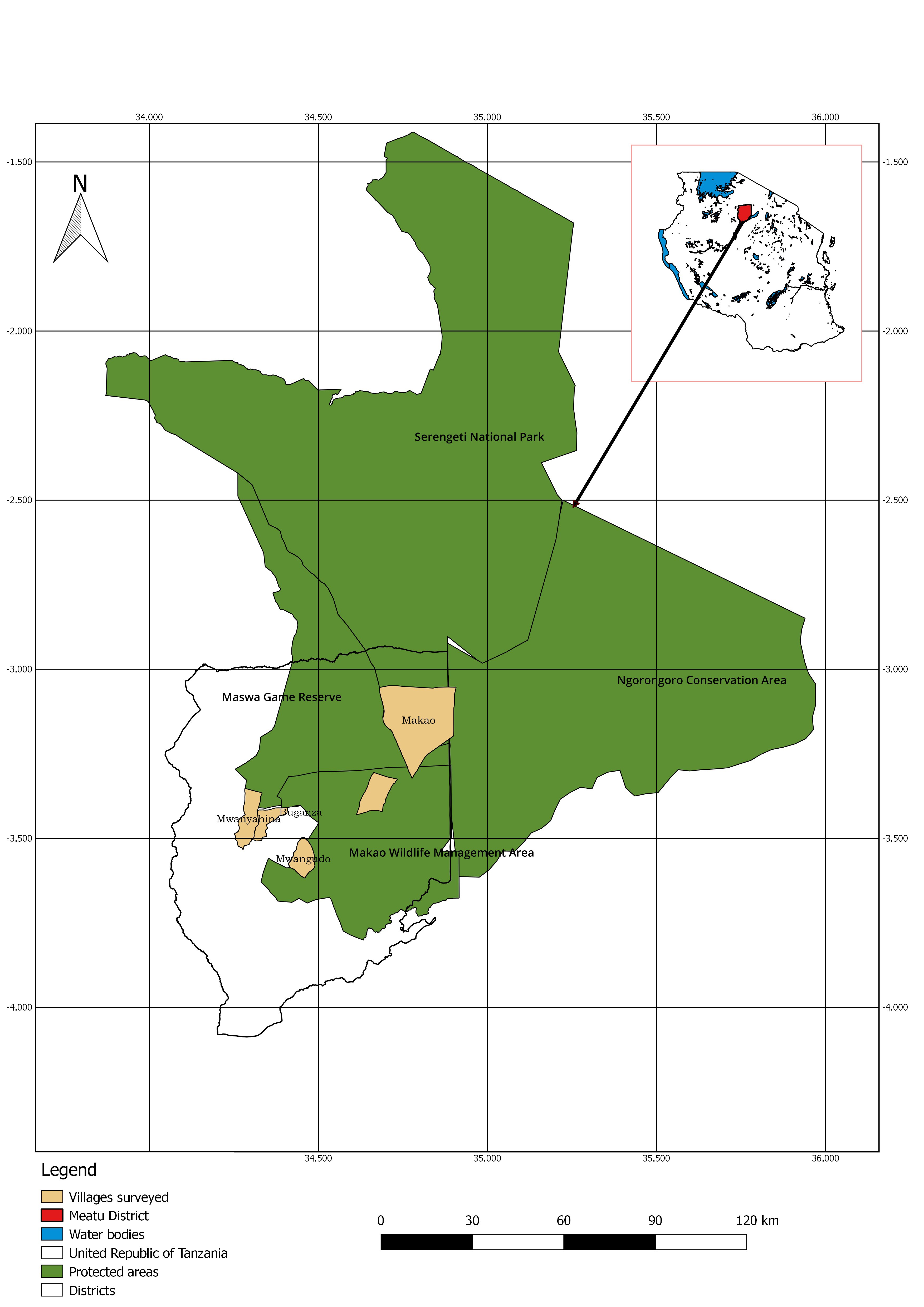
Study area The study was conducted in Meatu district, which is located between longitude 34°8' and 34.49"E and between latitude 2°57' and 4.9"S. Specifically the study was conducted in four villages namely Mwanyahina, Buganza, Mwangudo and Makao (Figure 1).
Figure 1. A map showing the surveyed villages from which samples were drawn Meatu is one of five districts of Simiyu region. The district has the size of about 8 871 km2 and a population size of about 296 616 people according to the 2012 Tanzania National Census (URT, 2012). The district experiences a uni-modal rainfall, which usually starts from October to May. The average annual rainfall ranges from 400 mm to 900 mm. Its vegetation is mainly composed of open bush savannah dominated by acacia species. The district borders Maswa Game reserve to the North (two-third of its size is in the district) and Ngorongoro Conservation Area (NCA) on the north eastern part. Furthermore, there are active wildlife conservation initiatives (Wildlife Management Areas) in some of the villages adjacent to the protected parks. The main economic activities of people include crop production, livestock keeping, bee keeping and mining. Sample collection Tsetse fly cluster samples (n=250) composed of three species: Glossina pallidipes (n=171), G. swynnertoni (n=58) and G. morsitans (n=21) and 100 cattle buffy coats samples from the four villages were used. Detailed method on collection and preservation of the flies and buffy coats samples have been described in Malulu et al. (2017). In brief, tsetse flies were sampled by Phenol and Acetone baited NZI, NGU, S3 and Biconical tsetse traps. Species and sex of the trapped tsetse flies were determined by taxonomic key as described in FAO (1982) tsetse manual. The flies were preserved in absolute ethanol contained Eppendorf tubes (each tube contained five tsetse flies of the same sex and species). Buffy coats were obtained in accordance to Murray et al. (1977) protocol and preserved on Whatman FTA cards (GE Healthcare, UK). DNA extraction Deoxy-ribose nucleic acid (DNA) extraction and PCR analysis were conducted at Vector and Vector Borne Diseases Institute, Molecular Laboratory, Tanga, Tanzania. The DNA from whole tsetse flies was extracted by ammonium acetate precipitation (Bruford et al., 1988). Absolute ethanol was poured off from each Eppendorf tube and sample ground using sterile pestle after being air dried overnight under room temperature. Phosphate Buffered Saline (PBS) (Ambion, USA) 500µL was added in each sample for homogenisation and from which 100µL of the supernatant was used for DNA extraction. Thereafter, 250 µL of Digisol buffer and 10µL of proteinase K (Qiagen GmbH, Germany) (stored at -20 0C) were added to the sample and vortexed for 30 secs and incubated at 55.0 oC for 1 h. Afterwards 300µL of ammonium acetate (Ambion, USA) was added to each Eppendorf tube and vortexed for 30 s repeatedly for 5 min then supernatant was transferred to new and sterile Eppendorf tubes. Then 1mL of 100% ethanol was added to the supernatant, vortexed and inverted gently 10-20 times for DNA precipitation followed by centrifugation at 21952g for 15 min. Supernatant was poured and pipetted off and 900µL of 70% cold ethanol was then added and the Eppendorf tubes inverted gently 10 times to rinse the DNA pellet. The DNA pellet was air dried overnight at room temperature then eluted in 100µL of Tris-EDTA (Sigma Aldrich, UK) and stored at -20 °C until further analysis. Chelex resign (Sigma Aldrich, USA) 20% was used in the DNA extraction of buffy coat samples preserved on FTA cards as described by Ahmed et al. (2011) and Ruiz et al. (2015). In brief, 10-hole punches (1.2 mm) were taken from the FTA card matrix (GE Healthcare, UK) using Harris Micro-punch (Harris, USA) and placed into 1500µL sterile Eppendorf tubes. Two hundred microlitres of 20% Chelex resign was added and vortexed for 20 s to ensure the beads are distributed evenly then incubated at 55 oC for 1h. Thereafter, the samples were agitated and centrifuged at 21952g for 10 s followed by boiling for 10min. Supernatant of 100µL was transferred to a new sterile Eppendorf tube and stored at -20 0C until used for PCR. DNA amplification Primers and PCR conditions used were those described by Njiru et al. (2005). ITS1 amplification was carried out in 25μL reaction mixture containing 2μL DNA template, 9.5μL PCR grade water (Ambion, USA), 12.5μL Taq1× Master mix (New England Biolabs MO270L) and 0.5μL of each ITS1 primers (CF: 5’-CCGGAAGTTCACCGATATTG-3’ and BR: 5’-TTGCTGCGTTCTTCAACGAA-3’). PCR cycles were; initial denaturation step done at 94 °C for 30 s, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing step at 58 °C for 45 s, polymerization at 68 °C for 1 min and final elongation step at 68 °C for 5 min. PCR products were visualized in 1.5% (w/v) agarose gel stained with Ethidium bromide through the ultra Violet-Trans-illuminator. Positive control (Trypanosoma evansi DNA) kindly donated by Dr. Hamid Ibrahim Noor (Khartoum University, North Sudan) was used and PCR water as negative control. A sample was classified in a respective species if found within base pair as indicated in brackets; Trypanozoon (480 bp), T. congolense; T. congolense savannah (700 bp), T. congolense forest (710 bp), T. congolense Kilifi (620 bp), T. simiae Tsavo (370 bp), T. simiae (400 bp), T. godfreyi (300 bp) and T. vivax (250 bp). Data analysis Data on tsetse species, cattle age group, sex, status of trypanosome presence and village Microsoft Excel sheets. Occurrence of trypanosomes infections in tsetse and cattle was calculated by determining total positives out of the total number of samples. Chi-square (𝜒 2) test was used further to compare trypanosome infections between cattle groups, villages and tsetse species using Epi info statistical software. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Results |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
PCR detection of Trypanosoma species Table 1. Occurrence of trypanosomes in cattle and tsetse flies based on ITS1 PCR
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
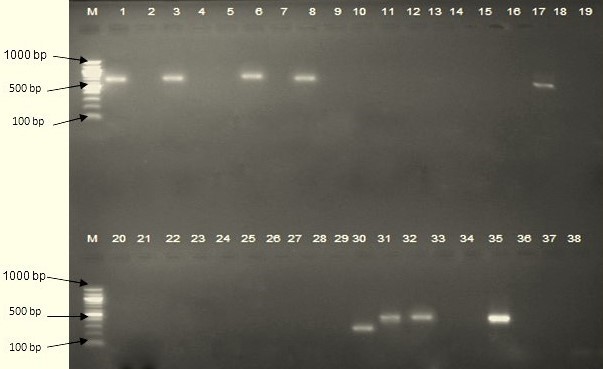
There was no significant difference in trypanosome infections between villages and between cattle groups (P> 0.05). However, infection in bulls (17.1%) was higher compared to other cattle groups. The occurrence of trypanosomes infections in tsetse flies was different between villages (P= 0. 03) all trypanosomes were from G. pallidipes of Buganza village. There was no significant difference in infections between Glossina species (P= 0.49). Fifteen positive samples from cattle were identified as T. congolense and produced fragments with molecular weights of between 600-700 bp (figure 2, lanes 1, 3, 6, 8, and 17). In tsetse flies three samples were identified as T. godfreyi (lane 30) and T. simiae (lanes 31 and 32) which produced molecular weight of 300 and 400 bp respectively. Sleeping sickness pathogens and co-infections were neither identified in tsetse flies nor in cattle.
Figure 2. Identification of Trypanosomes by ITS1 PCR, lane M- 1000 bp DNA ladder, lane 1-34 DNA, lane 35 positive control. The lanes 1, 3, 6 and 17 indicates T. congolense from cattle samples. For tsetse samples lane 30 indicates T. godfreyi and lanes 31 and 32 T. simiae. Lane 36 indicates negative control. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Discussion |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Analysis of this study reports presence and circulation of only animal infective trypanosomes both in tsetse flies and cattle. Identified trypanosomes species were T. congolense in cattle and T. simiae and T. godfreyi in tsetse flies. The occurrence of animal trypanosomes in cattle was higher than of other studies conducted in nearby areas (Ruiz et al., 2015; Manangwa et al., 2016). Nevertheless, Trypanosoma vivax that was detected in cattle through a parasitological study reported by Malulu et al. (2017) was not detected. This could be due to genetic diversity of the species in Tanzania that the current primers could not amplify also reported by Adams et al. (2009). The trapping of the Stomoxys and Tabanus known mechanical transmitters during the entomological study signifies its circulation. There was data inequality in tsetse samples collected in each village for analysis, which might have affected interpretation of trypanosomes infections results of Glossina species. This was because most trapped flies were teneral/hungry also reported in Muturi et al. (2011). But fact that all infections were animal infective from G. pallidipes, importance of all other species in disease transmission should not be neglected. The identification animal infective trypanosomes also suggest the need of instituting tsetse control in order to cut the transmission cycle. Different options that are suitable for the district exist including pour-on and insecticide treated targets. This is because the district experiences longer drought season than rain and water is a scarce resource. During drought it nearly impossible to practice either dipping or spraying. This will relieve the costs farmers are incurring in treating their livestock by reliant mainly on chemo treatment and the sleeping sickness risk they get exposed. Absence of human infective trypanosomes in cattle and tsetse has also been reported by Malele et al. (2011) and Ruiz et al. (2015) speculated this to be due to active treatment of cattle with trypanocidal drugs which kills human infective trypanosomes in livestock. The failure to identify human pathogen does not completely clear the doubts that there is no sleeping sickness in the area if protected areas continues to exist in which no tsetse control is done. The observation of T. godfreyi and T. simiae in tsetse flies suggests that the vectors fed on wild pigs (warthog), which farmers were also mournful on the destruction they caused on their crops and they are known human infective trypanosome reservoirs (Kaare et al. 2007). |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Conclusion |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
The current study did not find T. brucei rhodesiense in tsetse and cattle. However African Animal Trypanosomosis pathogens were observed in tsetse flies and cattle. Presence of these pathogens poses constrains in the livestock production and therefore tsetse control and proper animal treatment are advised. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Acknowledgments |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
This research was supported by Commission for Science and Technology (COSTECH) through Grant No: CST/RA.56/1426/2013. The funding source had no influence in either research design, data collection, analysis, interpretation or the decision to submit the article for publication. Preliminary results were presented as a poster at the 5th Join HAT platform-EANETT Scientific meeting, Kampala, Uganda, 3rd-4th October 2018. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Conflict of Interests |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Authors confirm that there is no conflict of interest regarding publication of this paper |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
References |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Adams, E.R., Hamilton, P.B., Malele, I.I., Gibson, W.C., 2008. The identification, diversity and prevalence of trypanosomes in field caught tsetse in Tanzania using ITS-1 primers and fluorescent fragment length barcoding. Infection, Genetics and Evolution, 8, 439-444. Adams, E.R., Hamilton, P.B., Rodrigues, A.C., Malele, I.I., Delespaux, V., Teixeira, M.M.G., Gibson, W., 2009. New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology 137, 641-650. Ahmed, A.H., Macleod, E.T., Hide, G., Welburn, S.C., Picozzi, K., 2011. The best practice for preparation of samples from FTA®cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasite and Vectors 4,1-7. Bruford, M.W., Hanotte, O., Brookfield, J.F.Y., Burke, T.A., 1988. Multilocus and single-locus DNA fingerprinting. Molecular Genetic Analysis of Populations 2, 287-336. Daffa, J., Byamungu, M., Nsengwa, G., Mwambembe, E., Mleche, W., 2013. Tsetse distribution in Tanzania: 2012 status. Tanzania Veterinary Journal 28, 12-20. FAO., 1982. Training Manual for Tsetse Control Personnel. Rome, Italy, Food and Agriculture Organization, pp. 247. Gibson W., 2003. Species Concepts for Trypanosomes: From morphological to molecular definitions?. Kinetoplastid biology and disease, pp. 1-6. Hoare, C.A., 1972. The Trypanosomes of Mammals. Oxford and Edinburgh, Blackwell Scientific Publications, pp.749. Ilemobade, A.A., 2009. Tsetse and trypanosomosis in Africa: The challenges, the opportunities. Onderstepoort Journal of Veterinary Research 76, 35-40. Kaare, M.T., Picozzi, K., Mlengeya, T., Fevre, E.M., Mellau, L.S., Mtambo, M.M., Cleaveland, S., Welburn, S.C., 2007. Sleeping sickness a re-emerging disease in the Serengeti?'. Travel medicine and infectious disease 5, 117-24. Lloyd, L., Johnson, W.B., 1924. The trypanosome infections of tsetse flies in Northern Nigeria and a new method of estimation. Bulletin of Entomology Research 14, 165-288. Malele, I.I., Magwisha, H.B., Nyingilili, H.S., Mamiro, K.A., Rukambile, E.J., Daffa, J.W., Lyaruu, E.A., Kapange, L.A., Kasilagila, G.K., Lwitiko, N.K., Msami, H.M., Kimbita, E.N., 2011. Multiple Trypanosoma infections are common amongst Glossina species in the new farming areas of Rufiji district, Tanzania. Parasite and Vectors 4, 1-8. Malulu, D.J., Kimbita, E., Tuntufye, H., Kinungh’i, S., Nyingilili, H., Mbilu, T., Kaboya, J., Lyaruu, E., Malele, I.I., 2017. An investigation on Glossina species and the prevalence of trypanosomosis in cattle in Meatu district, Tanzania. Journal of Parasitology and Vector Biology 9, 13-18. Manangwa, O., Ouma, J.O., Malele, I., Mramba, F., Msangi, A., Nkwengulila, G., 2016. Trypanosome prevalence in Glossina fuscipes fuscipes (tsetse) and cattle along the shores of Lake Victoria in Tanzania. Livestock Research for Rural Development 28. Murray, M., Murray, P.K., Mcintyre, W.I., 1977. An improved parasitological technique for the diagnosis of African trypanosomiasis. Trans R Soc Trop Med Hyg 71, 325-6. Muturi, C.N., Ouma, J.O., Malele, I.I., Ngure, R.M., Rutto, J.J., Mithöfer, K.M., Enyaru, J., Masiga, D.K., 2011. Tracking the Feeding Patterns of Tsetse Flies (Glossina Genus) by Analysis of Bloodmeals Using Mitochondrial Cytochromes Genes. PLOS ONE 6, 1-6. Njiru, Z.K., Constantine, C.C., Guya, S., Crowther, J., Kiragu, J.M., Thompson, R.C., Davila, A.M., 2005. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 95, 186-92. Ruiz, J.P., Nyingilili, H.S., Mbata, G.H., Malele, I.I., 2015. The role of domestic animals in the epidemiology of human african trypanosomiasis in Ngorongoro conservation area, Tanzania. Parasites and Vectors 8, 1-6. Sindato, C., Kibona, S.N., Nkya, G.M., Mbilu, T.J., Manga, C., Kaboya, J.S., Rawille, F., 2008. Challenges in the diagnosis and management of sleeping sickness in Tanzania: a case report. Tanzania Journal of Health Research10, 177-81. Steverding, D., 2008. The history of African trypanosomiasis. Parasites and Vectors, 1, 1-8. URT 2012. National population and housing census 2012. National Bureau of Statistics, United Republic of Tanzania, pp. 215. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|